The challenge: improve patient care using privacy-sensitive patient data
Hospitals collect fine-grained data about their patients to support medical treatment. This data is highly valuable to improve medical research, but also highly sensitive. The data is legitimately subject to strict regulations designed to protect patient privacy making it hard, long and sometimes impossible to be used for medical research.
In this blog post, we will show you how Sarus helps unlock patient data for research, without sacrificing patients’ privacy.
Dataset
In this demo, we are using a sample of an OMOP-like dataset which consists of 3 tables: patient information, type of treatments, and medication origins. These tables contain information linked to 1000 patients. This data is an extract from the CMS Synthetic Patient Data OMOP.
Getting access to such a fine-grained level of information on patients is notoriously difficult. It typically requires months-long data compliance processes, possibly with a formal approval from the data protection authority.
Find the data on our github.
Context for the case study
A researcher in heart diseases is given access to the dataset. They want to build a machine learning model to detect potential atrial fibrillation. The researcher will explore the data and build an adequate model.
Extract, explore, pre-process
Notebook intro
Find the notebook here.
The first lines of codes take care of importing the Sarus libraries and loading the data science and analytics API to be used and then connecting to the Sarus instance where the dataset was onboarded by the Data Administrator.
Explore the tables
Once the dataset has been selected, the researcher can browse its tables.

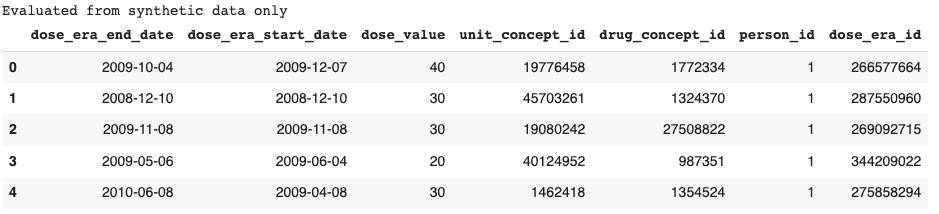
Notice the message: ‘Evaluated from synthetic data only.’ The privacy policy prevents all row-level information from being retrieved, so the application will return synthetic data instead. You can check out this article to learn more about our synthetic data generation model.
Synthetic data enables the researcher to explore the underlying data and get familiar with it even if they cannot see the real records.
Extract the relevant data with a SQL query and process them with pandas
After doing some exploration on the different tables from the dataset, the researcher defines the extraction of interest by using a SQL query.
Now that the relevant extract has been defined, the researcher will be able to define the preprocess pipeline necessary to train a Machine Learning model.
Then, the researcher builds a pipeline with scikit-learn, trains the model and checks its accuracy.
Here the researcher trained a RandomForestClassfieir using the scikit-learn library. Now, he will evaluate the model by comparing the accuracy score with the ratio in the data.
Note that the term ‘Whitelisted’ is in the output. It means that the computation was done using the original data without rewriting by exception (unlike computation executed against synthetic data or computation rewritten to have differential privacy guarantees). This comes from how the privacy policy was defined. It granted two exceptions to the researcher so that they can use cross_val_score and pipe.score. Thanks to the exceptions, the outputs of these methods can be retrieved by the researcher..
The accuracy of the model was close to 0.78, which means the detection rate was increased by more than 0.12 points. If the researcher considers the model to be accurate enough, it can be exported for future use. Or they can keep exploring new modeling strategies, still without spending any time waiting for data access approvals.
Conclusion
What were the benefits of using Sarus?
On the one hand, the hospital was able to make its data useful for research in a fully compliant way. The data was never copied or shared. The researcher never saw a single row of patient data. Every result was protected according to the privacy policy. .
On the other hand, the researcher was able to leverage the data right away. They could explore it freely thanks to synthetic data. Preprocessing and training of the machine learning algorithm were very standard; there was no need to learn a new way of coding.
Overall, the process was more secure and faster than it would be without using Sarus, and the robustness of the final machine learning algorithm is the same!
Want to give it a try? Book a demo!


.png)





